Single-cell Regulatory Occupancy Archive in Dementia
- 2 minsSingle-cell Regulatory Occupancy Archive in Dementia — scROAD
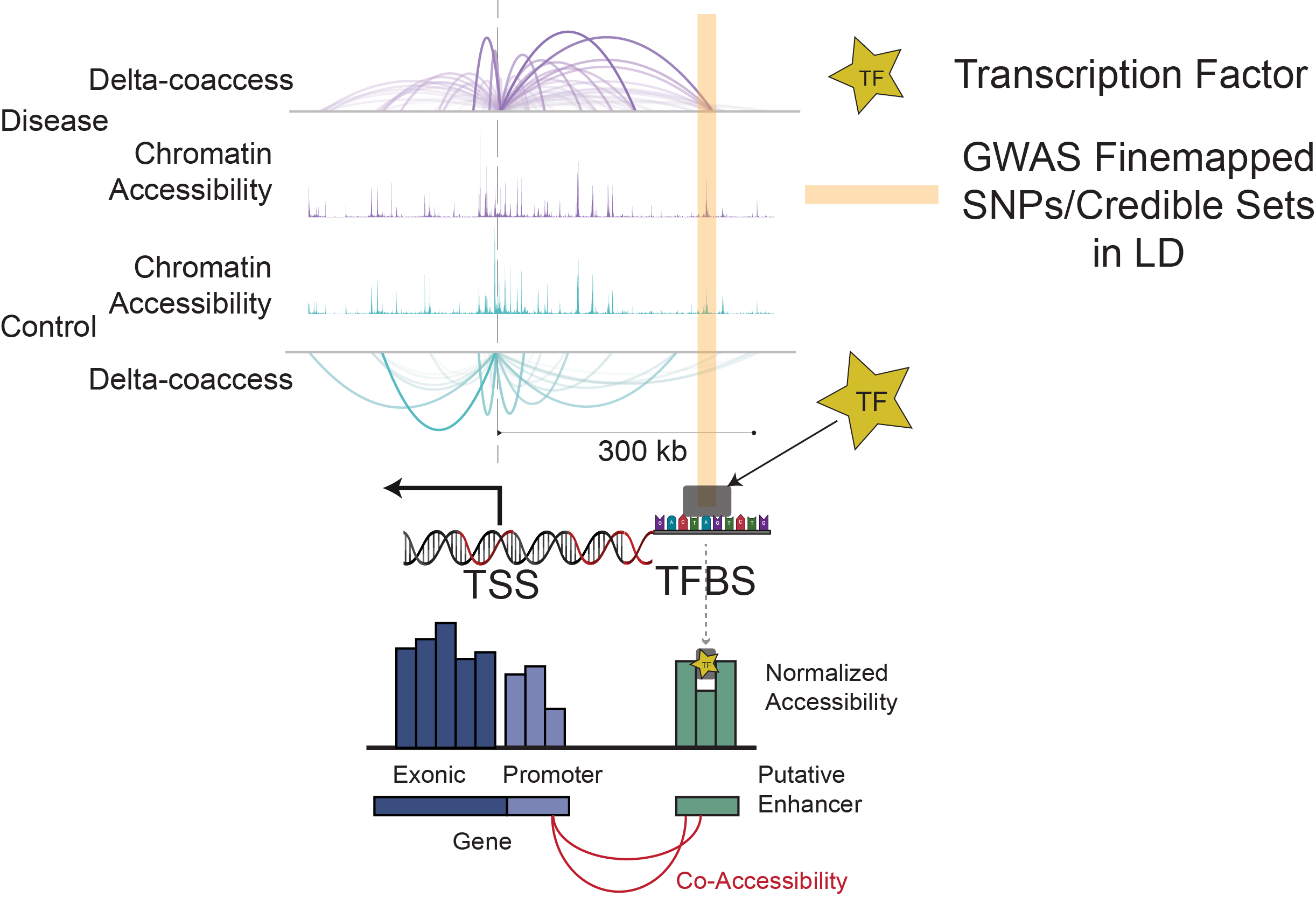
How can scATAC-seq data be utilized to study gene regulation and chromatin accessibility in diseases?
scATAC-seq data enables exploration of chromatin accessibility at a single-cell level, allowing identification of regulatory elements active in specific cell types. This technique is especially powerful for studying transcription factor binding sites (TFBS) and understanding gene regulatory networks in complex diseases like Alzheimer’s Disease (AD) and Pick’s Disease (PiD).
Our integrative framework incorporates genomic data from GWAS fine-mapping, snATAC-seq, and snRNA-seq to uncover genetic mechanisms driving neurodegeneration. By mapping disease-associated loci to specific cell types, we investigate functional alterations caused by genetic variants in AD and PiD.
I developed this interactive database, scROAD, which integrates:
- Single-cell chromatin data processed with Signac
- Cis-regulatory prediction from Cicero
- TF-binding occupancy inferred using TOBIAS
Co-accessibility networks generated by Cicero identify putative enhancer–promoter interactions. TOBIAS then quantifies condition-specific TF binding differences. Integrating these components into the scROAD interface enables users to explore transcription factor activity across disease and control states, providing a valuable resource for understanding regulatory changes in neurodegeneration.
About the Database
This database (Shi et al., 2025) provides comprehensive single-cell cCRE transcription factor occupancy information generated from snATAC-seq analysis of human postmortem prefrontal cortex (PFC). The dataset focuses on Alzheimer’s Disease and Pick’s Disease. For deeper details, please refer to the corresponding publication or contact the principal investigator.
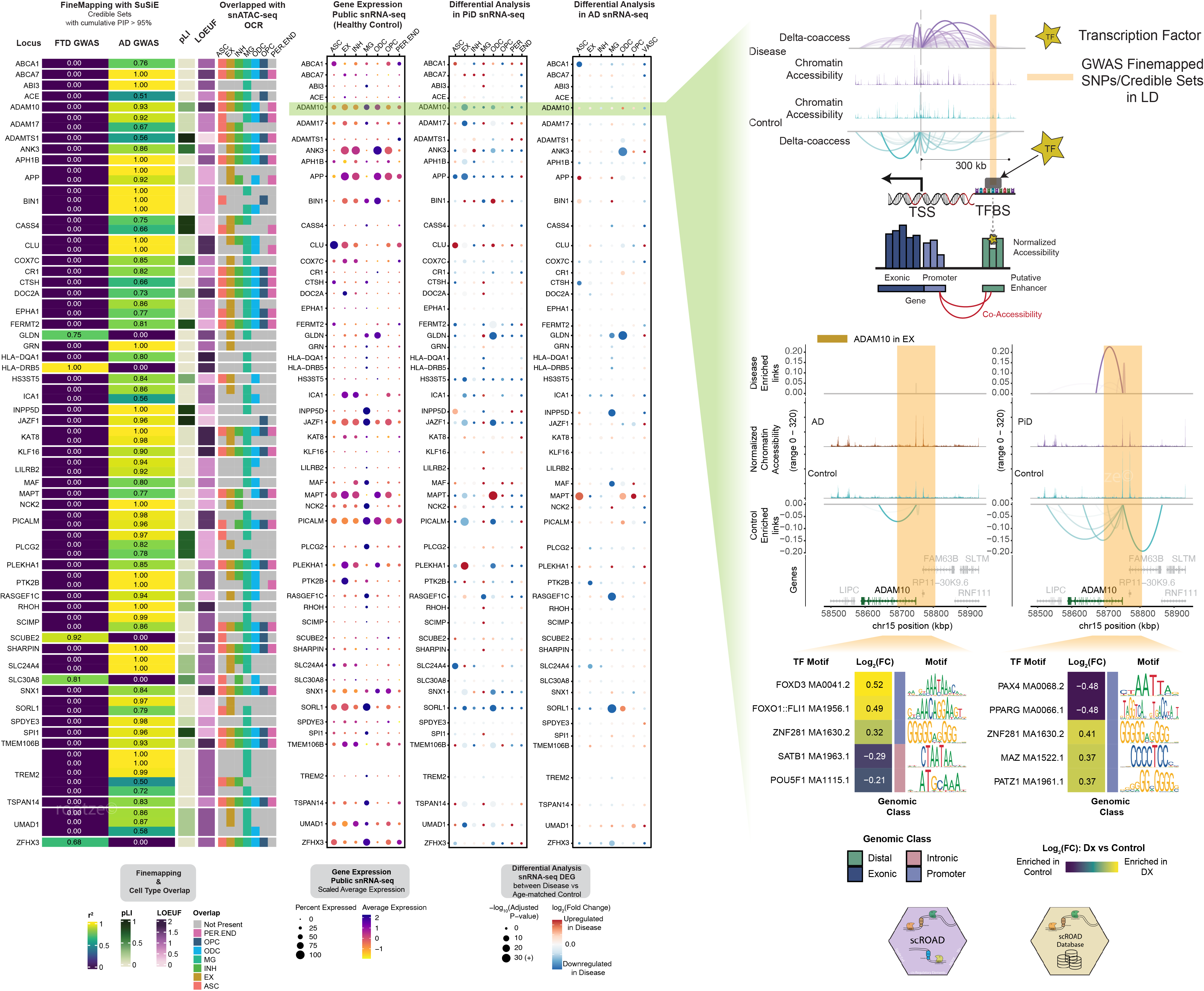 Fig.1 GWAS snATAC scCis-TF occupancy plot showcasing key transcription factor binding sites.
Fig.1 GWAS snATAC scCis-TF occupancy plot showcasing key transcription factor binding sites.
Running the code for Single-cell Regulatory Occupancy Analysis
The full Shiny application used to generate all TF occupancy visualizations in scROAD is available below:
➡️ View the full Shiny App code on GitHub
To run locally:
# Clone the repo
git clone https://github.com/rootze/scROAD.git
# Run the Shiny app
library(shiny)
runApp("shinyapp.R")