compact - Module Perturbation Analysis
Co-expression Module Perturbation Analysis in Cellular Transcriptomes
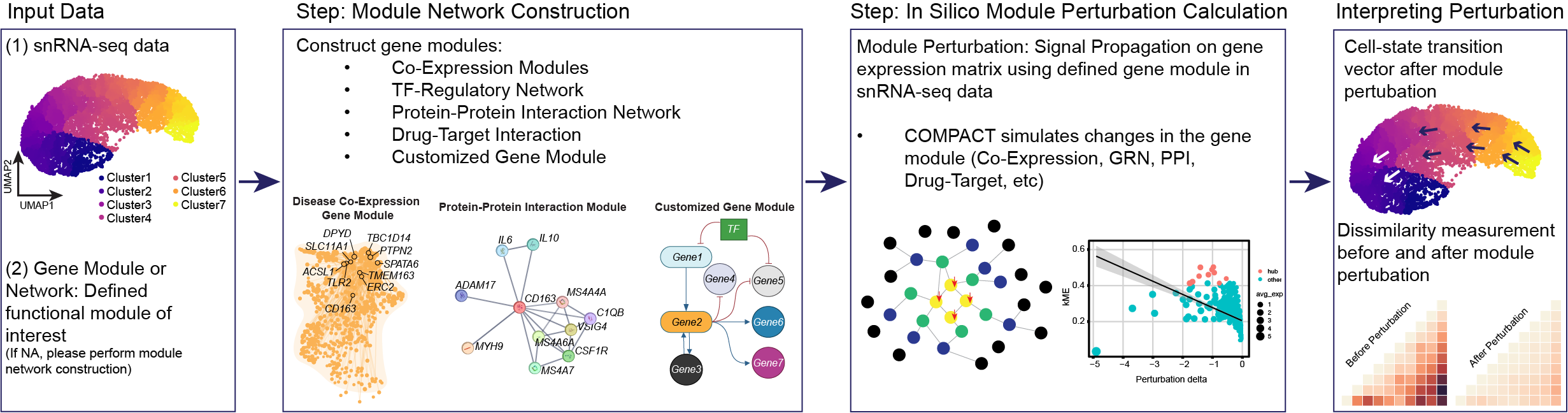
What We Can Do with functional annotated clusters in single-cell transcriptomes
Biological functions are governed by gene regulatory networks that orchestrate a diverse array of dynamic cell states. These networks are altered throughout development, aging, disease, and in response to molecular stimuli, however such perturbations are exceedingly difficult to measure directly with technologies like single-cell RNA-seq (scRNA-seq). Here we propose a novel computational framework, CO-expression Module Perturbation Analysis for Cellular Transcriptomes (compact), to perform in-silico gene expression perturbations in single-cell data, and to track downstream changes in cell state dynamics. Building off of our previous method high-dimensional Weighted Gene Co-expression Network Analysis (hdWGCNA), compact applies direct perturbations to co-expression network hub genes, and uses the network structure to propagate the perturbation signal to other linked genes. This framework is highly flexible to perform knock-in, knock-down, or knock-out perturbations on different networks and sets of genes and in different cell lineages, allowing researchers to explore a wide range of strategies mimicking various experimental conditions and interventions. We demonstrate the efficacy of compact across established paradigms of cellular dynamic response, including mouse oligodendrocyte differentiation, human disease-associated microglia, and existing single-cell perturbation data. Notably, our in-silico perturbation simulations with compact have unveiled disease-relevant phenotypic outcomes resulting from co-expression module perturbations in human disease-associated microglia. These findings offer valuable insights into the molecular mechanisms underlying cellular states in disease, and identify novel targets for therapeutic intervention. In summary, our work introduces compact as a powerful tool for functionally dissecting gene network perturbations and elucidating their disease-relevant impact. By leveraging the wealth of information contained within single-cell transcriptomic data, compact facilitates comprehensive analyses of gene regulatory networks, paving the way for advancements in our understanding of cellular dynamics and disease pathology.
Installation
We recommend creating an R conda environment environment for compact.
conda create -n compact -c conda-forge r-base r-essentials
conda activate compact
Install the required packages in R
# install BiocManager
install.packages("BiocManager")
BiocManager::install()
# install latest version of Seurat from CRAN
install.packages("Seurat")
# install devtools
BiocManager::install("devtools")
# install hdWGCNA, velocyto.R, and compact from devtools
devtools::install_github('smorabit/hdWGCNA', ref='dev')
devtools::install_github("velocyto-team/velocyto.R")
devtools::install_github('smorabit/compact')
Basic Tutorial
Source: Basic Tutorial
The basic tutorial covers the basics of using compact to perform module perturbation analysis on co-expression network/module on single-cell data.
This tutorial covers the basics of using compact to perform in-silico pertubation experiments in single-cell transcriptomics data. For this tutorial we will use an integrated single-nucleus RNA-seq dataset of microglia from three different studies of Alzheimer’s disease, as we described previously in the hdWGCNA paper.
The in-silico perturbations that we demonstrate in this tutorial are based on gene co-expression modules, which are unbiased clusters of genes that are highly co-expressed in a gene-gene co-expression network. In order to run these pertubations, we first need to perform co-expression network analysis and group genes into modules by running hdWGCNA. This has already been run on the tutorial microglia dataset, where we have identified four gene modules.
Conclusion : We used compact to demonstrate the basics of the compact method to apply an in-silico knockdown of a the microglial activation module M4. By down-regulating this module, the method predicted that cells would shift to a more homeostatic-like state.